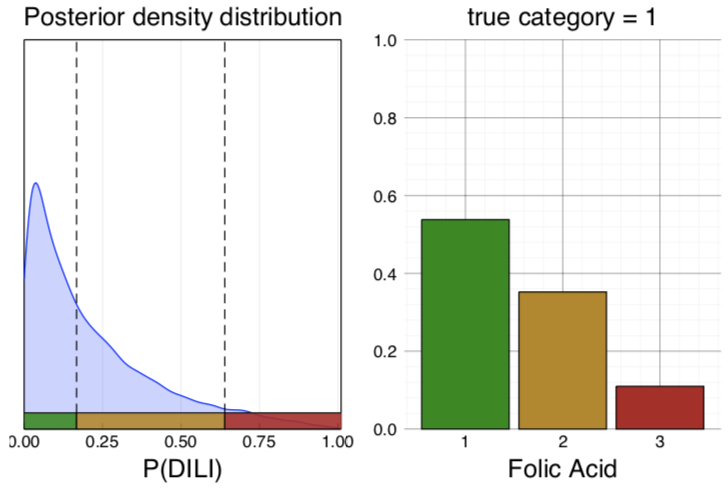
Predicting the toxicity of a drug preclinically reduces development costs and increases patient safety. Prediction models can be built using in vitro assays and physicochemical properties of compounds as features. AstraZeneca and Prioris.ai have developed a Bayesian neural network (BNN) to predict drug-induced liver injury.
Deep neural networks (DNNs) are used to predict toxicity for both a single target and multiple targets. Neural networks are popular due to their flexibility but are prone to overfitting and do not capture uncertainty. This might lead to overconfident predictions even when they are erroneous.
Bayesian neural networks (BNNs) use the architecture of neural networks and additionally describe each weight with a distribution, rather than a point estimate. The prior distributions used by BNNs regularize the weights and prevent overfitting. Furthermore, BNNs report the uncertainty in the predictions.
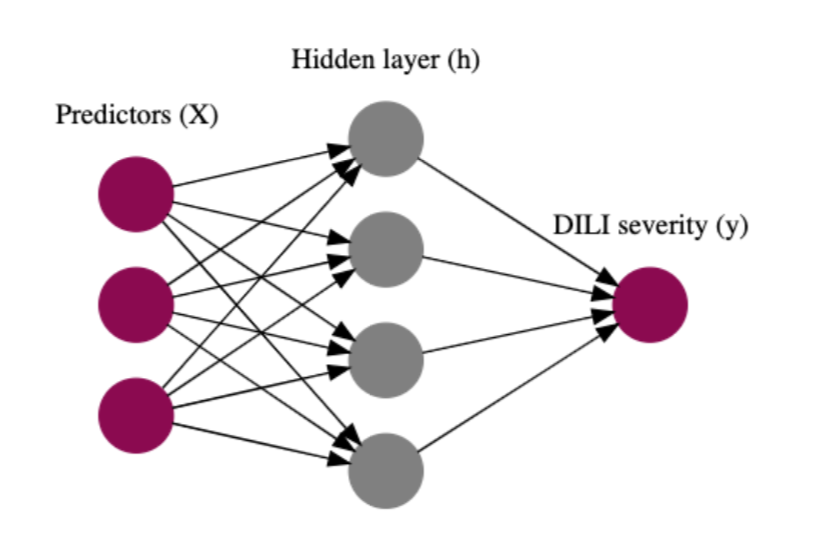
By separating the model and inference from each other, modern probabilistic programming languages (PPLs) make inference for Bayesian models straightforward. They provide an intuitive syntax to define a model, and a set of sampling algorithms to run the inference. Hence, only a generative model needs to be defined by a user to estimate parameters and make predictions.
In the context of probabilistic modelling, Julia provides a fertile ground: several native PPLs have been recently developed - Turing, Gen, Stheno, SOSS, Omega, indicating strong interest of the research community in the topic. The current model was implemented in Turing – a PPL embedded in Julia.
The original article is available here and the model can be run live at here via 'launch binder'.